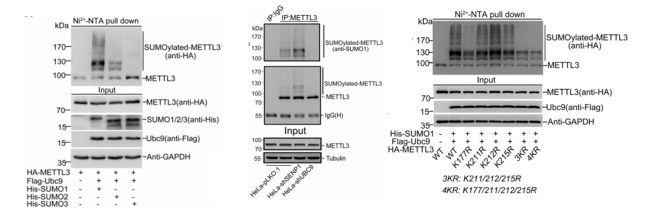
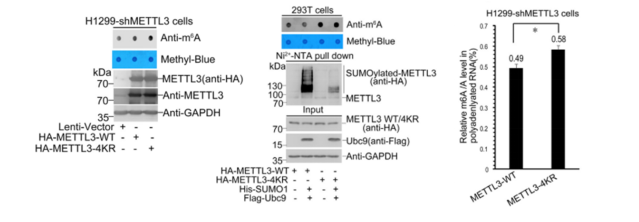
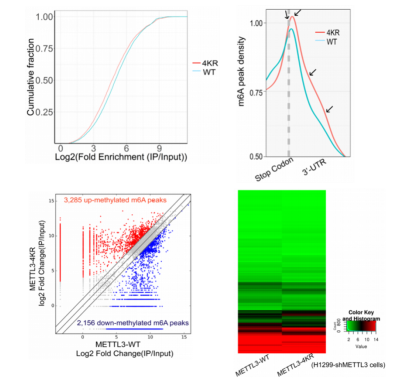
RNA methylation is currently the hottest area of ​​research. In the past three months, the number of articles with a factor of 10 or more in this direction has been close to 20. Yunxu Bio has conducted an in-depth analysis of RNA methylation research methods and ideas. Interested teachers can browse the pre -population of cloud pre-organisms (2018 National Natural Hotspot 2: Deep Analysis of RNA Methylation Research) . A partial list of high score articles in the past three months: On February 28th, another article on high-methylation of RNA methylation was published. Yujian Xiu Group Shanghai Jiaotong University School of Medicine in prestigious journals, "Nucleic Acids Research" (Impact Factor: 10.162) published a report entitled "SUMOylation of the m6A-RNA methyltransferase METTL3 modulates its function" article (cloud biological sequence provides a full set of RNA methylation Sequencing technology services). It is the first time to reveal a novel molecular mechanism by which SUMOylation regulates the m6A RNA methylase METTL3 and its catalytic function. A topic design with a good score of 10 or more will always be a step forward. When we are still trying our best to find the current hot m6A RNA methylation modification enzyme, the big cattle group has begun to study the translation of these enzymes themselves. Modifications, and the impact of the presence of these modifications on RNA methylation, have brought us a different RNA methylation feast: Research ideas: The Yu Jianxiu team identified the SUMO modification of METTL3 and its regulatory functions. They found that knocking down the SUMO-specific protease SENP1 in cells significantly reduced the level of m6A modification in intracellular RNA. This modification did not affect the stability of the protein and its localization in the cell, nor did it alter the interaction between the protein and METTL14 and WTAP. Through in vivo and in vitro experiments, they found that SUMOylation of METTL3 significantly inhibited the activity of m6A methyltransferase, promoted the formation of specific cancer cells and tumorigenesis. After stable knockout of METTL3, combined with m6A RNA methylation sequencing (MeRIP-seq) and second-generation high-throughput sequencing (RNA-seq), m6A RNA methylation map and downstream mRNA expression profiles were significantly changed. Since then, the argument that METTL3 regulates the modification and expression of downstream mRNA m6A by means of SUMOylation has been fully established. Study 1: METTL3 SUMO modification in vivo and in vitro identification The authors found that METTL3 was mainly modified by SUMO1 by Ni+-NTA Pull Down. After treatment with SUMO hydrolase Senp1, it was found that the METTL3 SUMO modification level did decrease. Endogenous expression of METTL3 also interacts specifically with SUMO1, and significant SUMOylation occurs. The authors then used the drug to stimulate the cells and found that the level of METTL3 SUMO in the body changed significantly. In vitro experiments confirmed interactions between the two. The authors supplemented the Co-IP experiment and confirmed that METTL3 SUMO levels were regulated by SUMO1 in vivo. To identify the METTL3 modification site in vivo, the authors constructed a series of METTL3 KR mutants, and finally identified that METTK3K177/211/212/215 is a SUMOylated substrate modification site. Study 2: METTL3 SUMO modification Protein SUMO modification is related to its biological processes such as localization, stability and protein interaction. The authors subsequently verified the physiological functions of METTL3 SUMO modification in the above three directions. First, the authors found through the Co-IP experiment that METTL3 SUMOization does not affect the stability of the protein itself. The authors subsequently ruled out the effect of METTL3 SUMO on its cellular localization. Finally, the authors found that the change in the degree of METTL3 SUMO does not affect its interaction with METTL14 or WTAP. So far, the three classical mechanisms of SUMOization have been eliminated. Then, based on the knowledge that METTL3 has RNA methyltransferase activity, METTL3 SUMO is highly likely to affect its enzymatic activity. Fortunately, the authors found that the expression level of m6A methylation in cell mRNA was significantly increased after expression of METTL3, but the level of methylation of m6A in intracellular mRNA was significantly decreased when the level of METTL3 SUMO modification was simultaneously increased. This part of the data advantageously confirms that METTL3 SUMOylation primarily affects its m6A RNA methyltransferase activity. Study 3: METTL3 SUMO-mediated transcriptome regulation The authors found that the size of the tumor was negatively correlated with the level of m6A modification in vivo by ectopic tumor formation experiments. The authors supplemented METTL3-WT (normal SUMOylation) and METTL3-4KR (non-SUMO) cell line m6A RNA methylation sequencing (MeRIP-seq) and transcriptome sequencing (RNA-seq) analysis, the results show that METTL3-4KR cell line The overall level of m6A modification increased, especially at the 3' UTR near the Stop Codon region. Transcriptome sequencing data showed that the expression levels of a large number of transcripts were significantly changed after METTL3-WT and METTL3-4KR were expressed in METTL3 knockout cell lines, respectively. After combining two sets of high-throughput data, it was found that 90 genes were significantly altered in both m6A RNA methylation level and transcript expression. The above experiments confirmed that METTL3 SUMOization will reduce the level of mRNA m6A modification, thereby affecting the expression level of the transcriptome and ultimately promoting tumor formation. to sum up: This study was the first to discover the presence of a protein modification of the m6A RNA methylation-modified catalytic enzyme. The team also believes that these key proteins must have other protein post-translational modifications (PTMs) that need to be further discovered. These PTMs include ubiquitination, phosphorylation, methylation, and acetylation. In the review of the paper, the paper has been highly recognized by the review experts. One of the referee pointed out that this research brings new elements to the regulation of m6A modification and answers the scientific question of how METTL3 self-regulates ("This paper brings new element insight METTL3 SUMOylation and m6A modification regulation, answering question of METTL3 self-regulation"); Interestingly, another referee also specifically mentioned the significant contribution of Yu Jianxiu Lab in the field of SUMO research ("Just one more comment. I obviously Am aware that the lab has made significant contributions to the SUMO field"). Researcher Yu Jianxiu of the Department of Biochemistry and Cellular Molecular Biology, School of Medicine, Shanghai Jiaotong University, and Dr. Zhao Wei, co-author of the paper, Du Yuzhen, a master student, and Hou Guofang, a joint doctoral student, are the co-first authors of the paper. Professor Chen Guoqiang and Professor Cheng Jinke also participated. The study. In addition, the research also received great help from researchers in the University of Zhejiang Liu Jianwei to identify m6A in mass spectrometry. Yunxu Bio is honored to participate in this wonderful research, undertaking m6A RNA methylation sequencing work and data analysis. As one of the earliest research service units in China to carry out RNA methylation sequencing services, Yunxun exclusively provides comprehensive m6A, m1A, m5C RNA methylation sequencing technology services, and is committed to escorting the research work of teachers. About the Author: Yu Jianxiu , male, Ph.D., is currently a Distinguished Professor and Doctoral Supervisor at the School of Medicine, Shanghai Jiaotong University. In 1990, he obtained a bachelor's degree from South China Agricultural University. In 1996 and 2000, he obtained a master's degree and a doctorate in zoology and molecular biology from Sun Yat-Sen University. Since September 2009, he has been a PI, Distinguished Professor of the Department of Biochemistry and Molecular Cytology, School of Medicine, Shanghai Jiaotong University, and a Distinguished Research Fellow at the State Key Laboratory of Oncogenes and Related Genes. He has published more than 50 research papers in internationally renowned academic journals Nature Chemical Biology, Molecular Cell, Nature Communications, EMBO J, Nucleic Acids Research, etc. He has presided over more than 10 national natural science key projects, major research projects, face projects, and major projects of the Ministry of Science and Technology. He has served as the National Science and Technology Commission's “Major New Drug Creation†Science and Technology Major Special Acceptance Expert, National Natural Science Foundation of China. Members of the scientific field disciplinary review team (second-instance experts) and the National Natural Science Foundation of China, major project review and acceptance experts. Mainly devoted to (1) to explore the role of protein modification in tumorigenesis and its molecular mechanism, the first identified SUMO modification of PTEN, and found that it directly mediates PTEN membrane binding and inhibits tumorigenesis and development, and analyzed PTEN-PI3K-AKT The molecular mechanisms of pathways; (2) the mechanism by which protein modifications regulate the production and metabolism of non-coding RNAs, and a series of studies have revealed a non-coding RNA regulatory network and mode of action mediated by protein modification. For example, it was found for the first time that the efficiency of miRNA/siRNA action was controlled by the degree of SUMOylation of TARBP2. Whole transcriptome sequencing Circular RNA sequencing RIP circular RNA sequencing m6A circular RNA methylation sequencing M5c circular RNA methylation sequencing Shanghai Yunxu Biological Technology Co., Ltd. Shanghai Cloud-seq Biotech Co.,Ltd Address: Lane 1066, Qinzhou North Road, Caohejing High-tech Development Zone, Shanghai Phone: 021-64878766 Website: Email: < NINGBO MEDICAL EQUIPMENT CO.,LTD , https://www.techartmeds.com

References: Circular RNA expression alteration in exosomes from the brain extracellular space after traumatic brain injury in mice
Cloud order related recommendation 