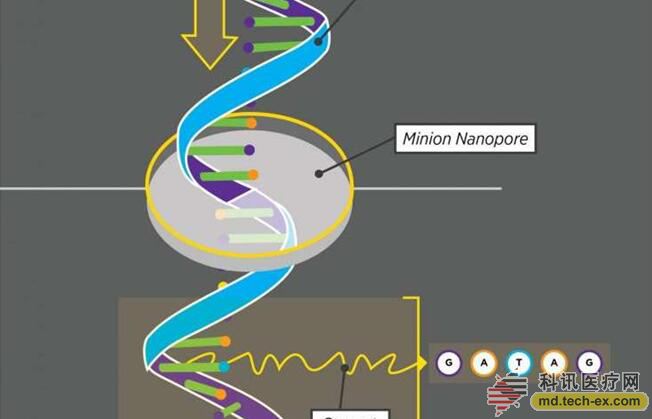
Release date: 2015-11-09 Researchers at Connecticut State University have sequenced RNA from the most complex genes known in nature by using a handheld sequencer that is smaller than a cell phone. When a DNA strand passes through the nanopore sequencer MinION, MinION can "see" five nucleotides. A set of five nucleotides generates a characteristic current that allows MinION to read the DNA code continuously. Source: Yesenia Carrero/Connecticut State University If DNA is a template for life, then RNA is the building contractor. RNA sequencing can help people interpret what is happening in cells. Brenton Graveley, a geneticist at the Institute of Systems Genomics at the Connecticut State University, his colleagues, Dr. Mohan Bolisetty, and graduate student Gopinath Rajadinakaran, and Oxford Nanoporous Technologies, claim that the company's MinION sequencer can sequence genes faster and better. And the cost is much lower than the technology that is commonly used today. They published their paper in the September 30 issue of Genomics Biology and introduced their sequencing technology. If your gene is a library and each gene is a book, then some genes can be read directly - but some genes are more like a "reader's choice plot" novel (a novel genre, provided by A number of possible episodes are available for the reader to choose, and then jump to the corresponding number of pages or paragraphs, so that the reader participates in the construction of the story). Researchers often want to know which version of the gene is expressed in the body, but this is not possible for complex "reader-selection plots" genes. Graveley, Bolisetty and Rajadinakaran tackled this problem in two ways. The first is to find a better gene sequencing technology. If the existing technology is used to sequence the gene, the researchers must first copy it using the same chemical process as our body, then cut the copied gene sequences into fragments, read each fragment, and then compare them. Each fragment finds out how they were stitched initially. This technique works because not every replicated gene sequence is cut into the same fragment. Imagine scrambling the sequence of shots in a movie. If you look at the same movie again, but the splices are slightly different, you can compare the two versions and find out which shots are connected. This technique is useless for the genes of the "practical plot", because if you copy them in the same way as the body, each replicated gene sequence is a bit different from the next one - or very large different. Different versions of this same gene are called gene subtypes. By splicing these different subtypes, it is impossible to accurately compare these fragments and find the original gene version. If the gene is a movie, "you may not know that lens 1 and lens 2 are synchronized," Bolisetty said. Just last year, this almost impossible thing suddenly became possible. The UK-based Oxford Nanopore has released a new nanopore sequencer and one for Graveley's lab. The nanosequence, called MinION, allows a DNA strand to pass through a tiny pore. The pores can only pass through five bases at a time - this is the "letter" that makes up our genes. There are four bases: G, A, T, and C, so there are 1024 possible five-base combinations, each of which produces a different current in the nanopore. GGGGA produces a different current than AGGGG, which produces a different current than CGGGG. By letting the DNA strands enter the nanopore and recording the signals they produce, the researchers know the sequence of the DNA. On another aspect of the solution, Graveley, Bolisetty and Rajadinakaran decided to play a big one. Instead of sequencing the old "practical plot" gene, they chose the most complex gene called Down's syndrome cell adhesion molecule 1 (Dscam1), which controls neural circuit connections in Drosophila brains. . Dscam1 has 38,016 possible subtypes, and each fruit fly can produce any one of them, but there are actually many subtypes that are still unknown. Dscam1 looks like this: X-12-X-48-X-33-X-2-X, X means always the same part, the number part may be different (the number itself represents how many different options there are) . In order to study how many different subtypes of Dscam1 exist in the fly brain, the researchers first had to convert the DNA molecule of the Dscam1 gene into RNA. If the DNA is a book or a instructions for use, RNA is a scribe who copies the contents of the book or instructions and translates it into protein. DNA contains the transcriptional pattern of all 38,016 subtypes in the Dscam1 gene, and each Dscam1 RNA reflects only one of the transcriptional methods. No one has previously used MinION to sequence RNA sequences, although this should be feasible, and demonstrating and demonstrating its effects will substantially advance the work in this area. Rajadinakaran extracted DNA from Drosophila brain, converted it to tNA, and isolated RNA fragments of Dscam1 DNA and passed them through a MinION nanopore sequencer. In one experiment, they not only found that 7899 of the 38,016 possible subtypes were expressed, but more subtypes might be expressed, if not all. “Many people say that 'MinION is of no use,'†Graveley said. “But our research shows that it can carry out research on the most complex genes known.†Studies have shown that gene sequencing technology can now be used more widely by researchers because MinION is relatively inexpensive and very light, with little room for experimentation. "This type of cutting-edge work will enable Connecticut State University to stay at the forefront of technology and increase the investment in our genomics research institute," said Marc Lalande, director of the Institute of Systems Genetics at Connecticut State University. "At the same time, it is also due to university academics. By investing in genomics at the University's Academic Plan, Brent Graveley can use his expertise to enable our faculty and students to successfully compete for donations and establish bioscience companies." Graveley will give a presentation on the study at the Oxford MiniION Nanopore Community Conference on December 3rd at the New York Genome Center. Brent Graveley's M inION gene sequencer uses advanced technology and is the size of an iPhone for just $1,000. Next, the researchers plan to do more in-depth research: thorough sequencing of each part of a single intracellular RNA, which cannot be done by traditional gene sequencing. "The magic is that this technology can change the way we study RNA science and the way we get information," Graveley said. "Plus MinION is a handheld sequencer that can be inserted into a notebook. It's incredibly cool. !" Source: Global Science Freeze Dried Fruits,Freeze Dried Fruit Bulk,Organic Freeze Dried Fruit,Freeze Dried Fruit Healthy Topower Technology Limited , https://www.topower-foods.com