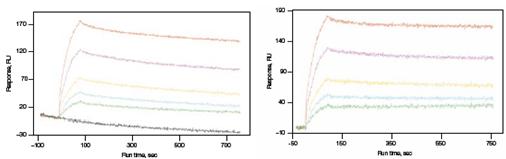
A new generation of parallel analysis of SPR biosensors: Surface plasmon resonance based biosensors are a non-labeled, real-time biomolecular interaction detection tool. Due to technical limitations, earlier SPR techniques used sequential analysis. Now, the latest generation of SPR instruments uses parallel analysis to measure the kinetics of the interaction of 1-6 pairs in a single injection. Compared with traditional techniques, parallel analysis has the advantages of good data quality, baseline linkage and high throughput. Another major benefit of parallel analysis is the baseline linkage, which makes the baseline smoother and the signal more accurate. For experiments in which the ligand is labeled by capture or there is a significant non-specific response, the baseline is uneven and the baseline drift levels are different after each regeneration and it is difficult to calibrate with a fixed calibration parameter. The parallel analysis method of the ProteOn system allows a channel to be buffered while flowing through the sample, and the baseline drift is observed. Then, the software can process the real-time calibration baseline to obtain a baseline stable and non-specific. The signal of the reaction. The figure below is an example of antibody screening. The anti-mouse IgG was first labeled on the chip to capture the monoclonal antibody in the culture supernatant, and then flowed through the antigen to measure the binding of the monoclonal antibody to the antigen. Because the monoclonal antibody and the antigen are bound, the monoclonal antibody itself is gradually dissociated from the IgG on the chip, so that the baseline is continuously reduced, and the rate of decrease is not uniform at different times; therefore, only through baseline linkage can we know what When the baseline is in the position, as shown in the left figure, the baseline is calibrated by software, as shown in the right figure below. If the conventional SPR sensor is repeatedly regenerated, there is no guarantee that the baseline drift at each injection is exactly the same, and naturally no accurate results are obtained. In summary, the new generation of SPR biosensors breaks through the traditional sequential analysis model, improves data quality, speeds up analysis, and reduces operating costs. It liberates the vast number of scientific research workers from the cumbersome labor of conditional exploration, and thus has been favored by researchers. [1] Rich RL and Myszka DG, Survey of the year 2004 commercial optical biosensor literature, J Mol Recognit 18, 431–478 (2005) [2] Rich RL and Myszka DG, Higher-throughput, label-free, real-time molecular interaction analysis, Anal Biochem 361, 1–6 (2007) [3] Tsafrir B., et al, The ProteOn XPR36TM Array System—High Throughput Kinetic Binding Analysis of Biomolecular Interactions, Cel. Mol. Bioengineering, Vol. 1, No. 4, December 2008, 216–228 HEBEI PINGLE FLOUR MACHINERY GROUP CO., LTD , https://www.plrollermill.com
Surface Plasmon Resonance (SPR) technology studies the interaction between biomolecules and small molecule drugs with non-labeling, real-time detection and small sample size, thus enabling basic life sciences, medical research and drug development. It is becoming more and more widely used and is almost an indispensable method for studying the interaction of biomolecules.
Biosensors based on SPR technology are only about 20 years old. According to the relevant literature of Professor David Myszka of the University of Utah, we can divide the development of SPR biosensor into 5 generations, the most advanced generation of sensors with high throughput and parallel analysis. Bio-Rad's latest ProteOn XPR36 protein interaction array system is one of the outstanding representatives. This article will focus on its technical characteristics and the improvement brought about by mutual research.
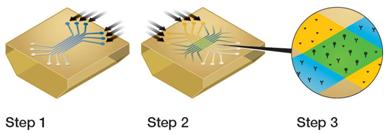
Like traditional SPR biosensors, the latest generation of technology also immobilizes one molecule on a chip, called a ligand, and another molecule flows through a ligand in a solution called an analyte. The difference is the way the two work. Take Bio-Rad's ProteOn XPR36 system as an example. It has a very unique flow cell called MCM (Multi-Channel Module). There are six parallel grooves on the MCM, which are firstly buckled on the surface of the chip in a vertical direction to form six parallel channels in the vertical direction for marking the ligand, as shown in the figure below. After the MCM is rotated by 90° and then the surface of the chip is bonded, six channels intersecting with the original are formed, and six different concentrations of analyte can be simultaneously flowed, as shown in the following figure “Step 2â€. Thus, the two sets of channels intersect to form 36 interworking sites, as shown in the following step "Step 3", which can achieve the kinetic constant (Kon, Koff) and dissociation equilibrium constant of the combination of it and up to 6 ligands. KD) is measured without the need to repeatedly regenerate the ligand. This type of analysis is called parallel analysis. Previously, no matter how many spots on the SPR sensor chip, only one concentration of analyte solution could flow through the chip at a time, called sequential analysis.
Many people intuitively believe that parallel analysis greatly increases throughput. This is certainly good, but in addition to drug screening by some pharmaceutical companies, for most laboratories, the biggest benefit of parallel analysis is that it facilitates the exploration of experimental conditions and improves data quality. The conventional SPR sensor uses sequential analysis to inject one concentration of analyte at a time. Between the two injections, it is necessary to elute all the analyte combined with the ligand with acid, alkali, high-concentration salt and the like, called regeneration. The exploration of regeneration conditions is often a very difficult task. Too mild reagents cannot be eluted cleanly, and strong reagents can easily damage the ligand, so that the final kinetic data deviates from the actual, and sometimes the deviation is still very obvious. Up to 100 times more.