Ningbo DOKEE Medical Technology Co., Ltd. , https://www.dokeemedical.com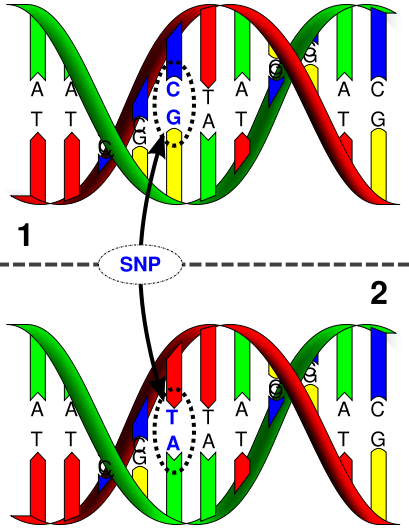
SNP (Single nucleotide polymorphism) is a single nucleotide polymorphism, which is a point mutation caused by the conversion, transversion, insertion or deletion of a single base at a specific nucleotide position of genomic DNA. There is a difference between individuals.
There are many methods for detecting SNPs, which are mainly based on the following four basic principles: (1) allele-specific hybridization; (2) endonuclease digestion; (3) primer extension; (4) oligonucleotide ligation reaction. In addition to well-established technologies such as gene sequencing and single-strand conformation polymorphism (SSCP), some newly developed gene chip technologies, denaturing high performance liquid chromatography (DHPLC), mass spectrometry, PCR and new PCR-based technologies Etc. also gradually entered this field and became the main means of detection. Due to the large number, wide distribution and stable inheritance of SNPs, many automatic operation techniques have been developed, and the analysis results can be processed by computer. Therefore, the application of SNP analysis technology has great potential in new drug research. It has significant industrial value in the fields of individualized medicine, clinical testing and molecular diagnosis.
SNP was originally used in medical research. With the simplification of its understanding and detection methods, SNPs have gradually penetrated into many fields such as genetics, ecology, crop breeding, etc., and the research objects have also expanded from human to important economic value. Research on the value of animals and plants, microorganisms, etc. It is believed that with the deepening of research, SNP will also have an immeasurable effect on population genetics, pharmaceutical industry, forensic science, and the evolution and improvement of species.
With the development of biotechnology and advances in technology, the application of SNP typing technology is becoming more extensive and open, such as:
1, SNP mapping
The International SNP Mapping Working Group outlined a map of 1.42 million SNPs with an average density of 1 SNP per 1900 base pairs. This map combines SNPs with human genome sequence, physical and genetic maps for further research on sequence variation, disease-associated genes, ethnic inheritance and genome scanning, and will provide insight into disease, diagnostic methods, and novel and effective methods. The development of treatment methods has far-reaching effects.
In order to meet the needs of China's human genome polymorphism program and research, China established the first human genome database in 2001, which is called “gene-oriented polymorphic databaseâ€, which mainly collects and studies the genome variation of Chinese population and its relationship with The relevance of complex diseases. At present, the relationship between hypertension, cancer and other diseases that seriously affect human health and even life and the corresponding allele SNP has been clarified. Due to the importance of SNP detection in post-genome planning, new technologies for high-throughput detection of SNPs are evolving, and high-level international journals such as Human Mutation, Genome Research, and Nucleic Acids Research have many questions about SNP Qualcomm. Report on quantitative detection technology.
According to the existing reports, the SNP detection method mainly focuses on comprehensive utilization of nanomaterial technology, multiplex PCR technology, various fluorescent probe design techniques and various fluorescent labeling techniques to improve the accuracy and throughput of detection, gene chip. And microsphere array technology also shows unique advantages in high-throughput detection of SNPs. In terms of detection technology, DNA arrays are undoubtedly the most ideal and have the most potential for development, but there are still some problems, such as high detection cost and insufficient reproducibility. Mass spectrometry is also widely used due to its high sensitivity and high accuracy. The use of nanomaterials and single-molecule detection technology is expected to achieve the ultimate goal of high-throughput detection of SNPs without PCR amplification.
With the advancement of technology and the completion of the sequencing of the genome of the species, the SNP database will be continuously enriched. Through all aspects of research, the SNP analysis technology will be more effective, and its application prospects will certainly be broader.
2. Association analysis of diseases
SNP-based association analysis found that genetic mutations in some complex diseases such as heart disease, cancer, diabetes, psychosis, etc. are a major challenge for human geneticists. Although these diseases expand in the family, they are usually inherited from generation to generation. As a result, the traditional family linkage analysis that has been successfully applied to Mendelian diseases is limited in this use, and complex diseases are the result of a combination of genetic and environmental effects.
At present, the most powerful method for discovering these polygenic diseases is to use statistically related DNA markers associated with disease genes to statistically compare the allele frequencies of the marker groups in the case group and the control group. SNP is very good for this analysis. A mark. Leonid Kruglyak used computer simulations to estimate the extent of the middle of the genome. He predicted that a genome-wide map containing 500,000 SNPs would be useful for identifying disease-causing genes for complex diseases. Two prostate cancer-related genes have been discovered through SNP association, and steroid hormones have been found to be associated with breast cancer and prostate cancer. With the continued discovery and application of SNPs, this method will be more widely used.
3. Drug design and application
Studies of SNP polymorphisms have shown a link between genetic polymorphisms and individual sensitivity or responsiveness to drugs, leading to a new discipline, Pharmacogenomics. It mainly studies the influence of genetic factors on the effects of drugs and the differences in drug response of different individuals, so as to select experimental populations for clinically targeted rational drug use and response to drugs according to different genotype individuals or groups, improve drug design, and market for drugs. Post-implementation monitoring provides a theoretical basis. After the drug is marketed, a drug monitoring system can be established to establish a SNP map of a susceptible individual by comparing the DNA of the subject with serious side effects and the control without the reaction, and by comparing the DNA of the well-infected individual with the non-active individual. , Establish a positive effect individual cSNP diagram. Together, they form a comprehensive drug response model that helps patients select effective therapies with fewer side effects.
4. Individual identification and parental identification
Although the SNP site provides relatively little information, the high density of the distribution compensates for the lack of information. Many SNPs have been used in forensic science, such as the polymarker system for paternity identification, and the probability of exclusion in yellows is 0.9962. SNP is expected to be widely used in forensic individual identification and paternity testing.
In recent years, due to the development of SNP typing technology, as a widely distributed and relatively stable genetic marker SNP, it has advantages such as easy to evaluate allele frequency and easy genotyping, which makes it subject to medical, biological, pharmaceutical and other fields. A wide attention has been paid. We believe that the in-depth study of SNP will definitely play a positive role in the research of human genetics, molecular genetics, pharmacogenetics, forensic science and the diagnosis and treatment of diseases.